Research suggests greater complexity in directing cargo within the cell
October 21, 2019
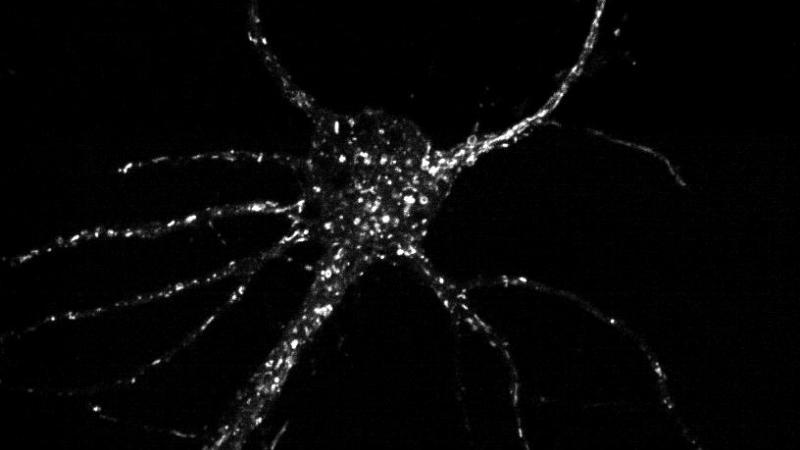
A new imaging technique that makes it possible to match motor proteins with the cargo they carry within a cell is upending a standard view of how cellular traffic reaches the correct destination. The research, which focuses on neurons and sheds light on some neurodegenerative diseases, was published in the current edition of the journal Traffic.
“What we see is that there must be some other regulation beyond what is known that is directing kinesin motor proteins within a cell,” said Marvin Bentley, assistant professor of biological sciences and member of the Center for Biotechnology and Interdisciplinary Studies at Rensselaer Polytechnic Institute. “There’s a mysterious interaction, and we don’t know what it is.”
All living cells are crisscrossed with a network of tiny filaments called microtubules, which serve as a cellular highway for the movement of nutrients, replacements parts, and other vital materials. Motor proteins, molecules resembling a pair of legs supporting a tall stalk-like tail, ferry cargo along the cellular highway. The human genome expresses 45 motor proteins of a type known as kinesins, of which 15-20 are involved in transporting vital parts in packages known as vesicles. But how do the kinesins know who to carry and where to go?
To answer similar questions, researchers often turn to expressing a target protein with a green fluorescent protein (GFP) tag attached to it. They can then expose a sample to ultraviolet light and observe the presence of tagged proteins. But simply GFP tagging motor proteins to see which ones go where doesn’t work – the ubiquity of the motor proteins as they go about their business floods the scene like a cellular version of light pollution.
As a workaround, previous research expressed only the legs – called the “motor domain” – tagged with GFP, and observed where the disembodied legs traveled. Those experiments were provided evidence in support of the “smart motor” theory, which posits that regulation is accomplished by the preference of the motor domain, with each kinesin traveling only on select microtubules and transporting vesicles belonging at the end of those microtubules.
But Bentley’s approach suggests the “smart motor” isn’t the only regulation at play.
Bentley’s lab developed a technique to express only the tail of each kinesin tagged with GFP. The tails bind to specific vesicles, showing which kinesin binds to which cargo, a different take on who carries what and where. For a few hours after the cell begins expressing kinesin tails – before the view is flooded with an increasing number of glowing tails – the researchers were able to observe all of the vesicles to which each kinesin binds.
The results show that when attached to vesicles, the “smart motor” theory doesn’t always hold true. Some kinesins don’t appear in the area indicated by their “smart motor” preference, while others appear in areas they shouldn’t even be able to reach.
“It’s clear that it’s not solely the preference of the motor domain that determines where these things go, there’s something else that’s regulating them,” said Bentley.
Bentley suspects that adapter proteins are involved, and hopes further research reveal the exact mechanism.
“The movement of essential neurotransmitters and metabolites within a cell is amazingly complex, but we must understand these fundamental processes before we can control diseases where this cellular traffic has gone awry, particularly in neurons,” said Curt Breneman, dean of the School of Science. “Marvin’s inventive approach to this problem has yielded a fascinating insight and a new direction for further research.”
Bentley was joined by researchers at the Oregon Health & Science University and Duke University. “A Novel Strategy to Visualize Vesicle-bound Kinesins Reveals the Diversity of Kinesin-mediated Transport” was supported by the National Institutes of Health.