Rensselaer engineers aim to make better biopharmaceuticals through deeper analysis
November 3, 2021
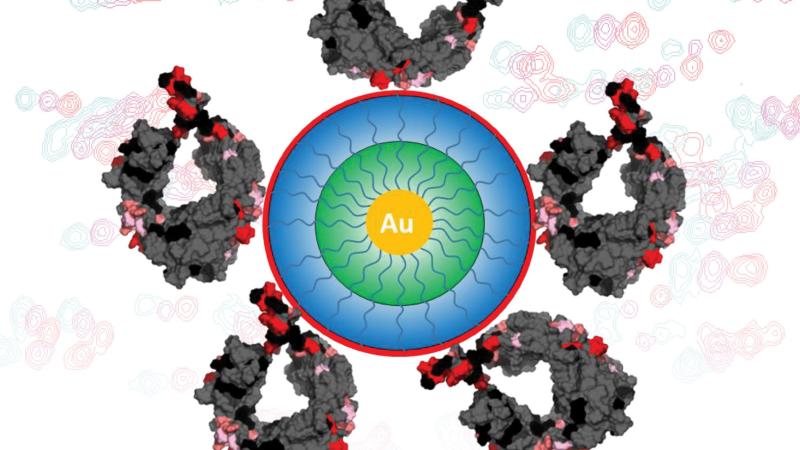
The process of purifying biopharmaceutical drugs remains a costly and time-consuming challenge. A deeper understanding of how unwanted elements within biomanufactured proteins bind to the molecules developed to remove them could help researchers make purity processes more efficient, more complex, and increasingly scalable.
In research published in Langmuir, a team led by Steven Cramer, an endowed chair professor of chemical and biological engineering at Rensselaer Polytechnic Institute, explored the fundamentals of how different molecules interact with various surfaces during the purification process.
Cramer is a leading expert in chromatographic bioprocessing, a separation technique used in biopharmaceutical purification to selectively choose which components of a protein mixture should be kept and which components should be removed. Ions or molecules, known as ligands, are developed to bind to specific components that should either be saved or discarded during this process.
“This is part of a very big effort to understand the fundamentals of how these molecules interact with surfaces,” said Cramer, a member of the Rensselaer Center for Biotechnology and Interdisciplinary Studies (CBIS), where the work was performed. “Our group is trying to ramp up the intellectual level of this kind of analysis in a variety of ways.”
This new paper builds upon research by the Cramer Lab recently published in Biotechnology and Bioengineering. In that work, researchers used nuclear magnetic resonance (NMR) spectroscopy and complex computer simulations to examine the fundamentals of how different molecules interact with various surfaces and ligands, including how and where binding happens, and if certain molecular interactions affect the binding process.
Working with Merck Pharmaceuticals and Bio-Rad Laboratories and using the NMR core facility in CBIS run by co-author Scott McCallum, the team looked at the Fc part of an IgG1 antibody and how ligands interact with that section of the antibody protein in particular. (If you were to picture an antibody looking like the letter “Y,” the Fc part would be the tail.) IgG1 antibodies are used in a large majority of biopharmaceutical drugs, meaning a deeper understanding of how their components interact with various molecules and ligands could have widespread implications.
In the Langmuir paper, the team took its exploration even deeper by adding nanoparticles to the surface of various proteins in order to see exactly where the ligands were binding.
“With this approach, we can then also look at some of the subtle interactions that are happening,” Cramer said. “Sometimes these ligands come together and form these clusters of ligands, and these clusters actually drastically change the behavior.”
This work, Cramer said, could lead to the development of new and improved materials and ligands. It also could help researchers develop more nuanced and specific ways of separating out unwanted molecules that are very similar to another type of molecule that needs to remain. All of these advancements could improve the drug purification process, making it more efficient and effective.
“Probing IgG1 FC–Multimodal Nanoparticle Interactions: A Combined Nuclear Magnetic Resonance and Molecular Dynamics Simulations Approach” was published October 11.